Search results
Search for "open source" in Full Text gives 20 result(s) in Beilstein Journal of Organic Chemistry.
Using the phospha-Michael reaction for making phosphonium phenolate zwitterions
Beilstein J. Org. Chem. 2024, 20, 41–51, doi:10.3762/bjoc.20.6
- displayed in Scheme 1. All simulations were performed with COPASI, an open-source software [48]. The second-order rate constants were obtained by fitting the experimental time traces until a fully consistent data set, being valid for all experimental conditions, was established. For X-ray structure analyses
NMRium: Teaching nuclear magnetic resonance spectra interpretation in an online platform
Beilstein J. Org. Chem. 2024, 20, 25–31, doi:10.3762/bjoc.20.4

- an open source web component [30][31]. It is derived from a freely available chemoinformatics tool set directed at scientific and educational applications [32][33]. It is a tool to display spectral data and metadata that has been developed as part of our efforts to aid better handling and evaluation
Synthesis and characterisation of new antimalarial fluorinated triazolopyrazine compounds
Beilstein J. Org. Chem. 2023, 19, 107–114, doi:10.3762/bjoc.19.11
- , Australia Discovery Biology, Griffith University, Nathan, QLD 4111, Australia NatureBank, Griffith University, Nathan, QLD 4111, Australia 10.3762/bjoc.19.11 Abstract Nine new fluorinated analogues were synthesised by late-stage functionalisation using Diversinate™ chemistry on the Open Source Malaria (OSM
- ; triazolopyrazine; scaffold; Open Source Malaria; Introduction Malaria is an infectious disease caused by Plasmodium parasites and is a major global threat to human health. The WHO World Malaria Report 2021, estimates 241 million cases of malaria and 627,000 deaths globally in 2020, an increase of 12% from the
- targets are urgently needed to combat the global problem of parasite drug resistance. For more than 10 years, the Open Source Malaria (OSM) consortium [4] has had an interest in identifying and developing novel antimalarial compounds that belong to a variety of chemotypes, one of which includes the 1,2,4
Synthesis and physicochemical evaluation of fluorinated lipopeptide precursors of ligands for microbubble targeting
Beilstein J. Org. Chem. 2021, 17, 511–518, doi:10.3762/bjoc.17.45

- preparation and upon time were determined on 5–10 slides using Fiji (an open-source image processing package [45]) and the standard deviations were calculated using Origin9 (OriginLab Corp. Northampton, MA, USA). Adsorption kinetics of perfluoroalkylated lipopeptides 1–3 and the hydrocarbon analog 4 at the
Semiautomated glycoproteomics data analysis workflow for maximized glycopeptide identification and reliable quantification
Beilstein J. Org. Chem. 2020, 16, 3038–3051, doi:10.3762/bjoc.16.253

- quantification in LaCyTools [16]. A python script was developed to facilitate this step (Supporting Information File 3). LaCyTools was chosen because it is open-source, can be applied for a large number of samples (thousands of samples in one study have been reported [19]), and allows data curation and
- clusters, MS1-based approaches show a highly complementary performance by identifying glycopeptides for which no MS/MS data is present. For the latter, GlycopeptideGraphMS is a highly valuable tool as it is easy to use, fast, and open source. Glycopeptide curation and quantification in LaCyTools Upon
A consensus-based and readable extension of Linear Code for Reaction Rules (LiCoRR)
Beilstein J. Org. Chem. 2020, 16, 2645–2662, doi:10.3762/bjoc.16.215

- demonstrating non-trivial functional-group (Table 5) relations between monosaccharides (Figure 2). We used RDKit, an open-source cheminformatics toolkit, to identify chiral centers and further determine stereochemical equivalence classes. Monosaccharides were clustered with an 80% stereo-similarity threshold
- of reactions possible, nor open-source software available to implement the parsing of such rules. Representing reaction rules in Linear Code is not easy because of a few ambiguous cases not completely described in the initial Linear Code paper. Subsequent studies, therefore, have developed their own
Computational tools for drawing, building and displaying carbohydrates: a visual guide
Beilstein J. Org. Chem. 2020, 16, 2448–2468, doi:10.3762/bjoc.16.199

- and GLYCOSCIENCES.de databases. DrawRINGS is an efficient tool for sketching glycan figures as well as translating to (and from) the KCF and IUPAC text formats. DrawGlycan-SNFG. DrawGlycan-SNFG [31] is an open-source program available with a web interface (Figure 7, top) at http
Clustering and curation of electropherograms: an efficient method for analyzing large cohorts of capillary electrophoresis glycomic profiles for bioprocessing operations
Beilstein J. Org. Chem. 2020, 16, 2087–2099, doi:10.3762/bjoc.16.176

- similar groups, making it easier to define peaks manually. Open-source software is then used to determine peak areas of the manually defined peaks. We successfully applied this semi-automated method to a dataset (containing 391 glycoprofiles) of monoclonal antibody biosimilars from a bioreactor
- other tested fully automated software. Briefly, the method performs clustering analysis of glycomic electropherograms to group them into manageable clusters, followed by subsequent quantitation after semi-automated curation using the open source software HappyTools [16]. The clustering and migration
- therefore describe a method whereby we perform clustering analysis of large cohorts of glycomic CE-LIF electropherograms, breaking them down into smaller, more manageable groups of similar electropherograms, followed by using open-source software for quantitation. The computational approach can be adapted
Antibacterial scalarane from Doriprismatica stellata nudibranchs (Gastropoda, Nudibranchia), egg ribbons, and their dietary sponge Spongia cf. agaricina (Demospongiae, Dictyoceratida)
Beilstein J. Org. Chem. 2020, 16, 1596–1605, doi:10.3762/bjoc.16.132
- 12-deacetoxy-4-demethyl-11,24-diacetoxy-3,4-methylenedeoxoscalarin in all three samples. Proposed relative configuration of 12-deacetoxy-4-demethyl-11,24-diacetoxy-3,4-methylenedeoxoscalarin. Selected NOE correlations are indicated with arrows. The model was obtained using Avogadro, an open-source
In silico rationalisation of selectivity and reactivity in Pd-catalysed C–H activation reactions
Beilstein J. Org. Chem. 2020, 16, 1465–1475, doi:10.3762/bjoc.16.122
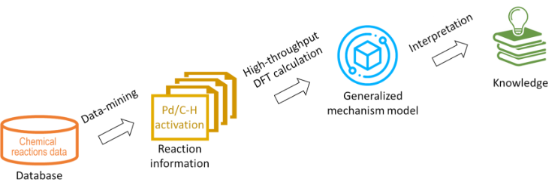
- computational data, a threshold to distinguish between two possible reaction mechanisms was established. Computational Methods The NWChem, an open source software package, was used for the DFT calculations. It is easily scalable and designed to solve large scientific computational problems efficiently employing
Anthelmintic drug discovery: target identification, screening methods and the role of open science
Beilstein J. Org. Chem. 2020, 16, 1203–1224, doi:10.3762/bjoc.16.105
- . Phenotypic screening remains important, and there has been much progress in open-source systems for compound screening with parasites, including motility assays but also high content assays with more detailed investigation of helminth physiology. Distributed open science compound screening programs, such as
- discovery is to reduce the cost, by introducing research strategies that make drug innovation more efficient [72]. Open source drug discovery is a model that seeks to completely open up the research process [78]. This has several radical advantages that challenge traditional drug discovery. Secrecy and the
- effective new drugs [79]. The latter is an open database for researchers to upload scientific data, including biological results, so that others may use it [80]. Perhaps the biggest example of open source drug discovery in its pure form is the Open Source Malaria project [81][82]. This is a platform for
A smart deoxyribozyme-based fluorescent sensor for in vitro detection of androgen receptor mRNA
Beilstein J. Org. Chem. 2020, 16, 1135–1141, doi:10.3762/bjoc.16.100

- resulting from the 60-AR_RNA cleavage. Prediction of the RNA secondary structure with the folding energy before and after cleavage was performed by MFold open source [21]. The SDFS functional activity. A–C) Emission spectra of the assembled SDFS (green line), T1–T5 chains mix without annealing (green dotted
Bacterial terpene biosynthesis: challenges and opportunities for pathway engineering
Beilstein J. Org. Chem. 2019, 15, 2889–2906, doi:10.3762/bjoc.15.283

- . antiSMASH [48] is the only open source web application that annotates bacterial terpene BGCs. As a result of the low conserved sequence similarity between TCs, terpene BGCs are missed more frequently by antiSMASH than other BGC classes [49]. In contrast to the well-characterized biosynthetic rules of
Low-budget 3D-printed equipment for continuous flow reactions
Beilstein J. Org. Chem. 2019, 15, 558–566, doi:10.3762/bjoc.15.50

- various screws are necessary for the assembly (Figure 5). For a full part list, CAD files of the printed parts and manufacturing details see the Supporting Information. The control command program was written on the open-source Arduino software and was fully adaptable to syringes from 1 mL to 50 mL. After
A recursive microfluidic platform to explore the emergence of chemical evolution
Beilstein J. Org. Chem. 2017, 13, 1702–1709, doi:10.3762/bjoc.13.164

- an automated recursive platform based on droplet microfluidics, could be used to induce artificial chemical evolution by iterations of growth, speciation, selection, and propagation. To explore this, we set about designing an open source prototype of a fully automated evolution machine, comprising
The digital code driven autonomous synthesis of ibuprofen automated in a 3D-printer-based robot
Beilstein J. Org. Chem. 2016, 12, 2776–2783, doi:10.3762/bjoc.12.276
- Philip J. Kitson Stefan Glatzel Leroy Cronin WestCHEM, School of Chemistry, The University of Glasgow, University Avenue, Glasgow G12 8QQ, UK 10.3762/bjoc.12.276 Abstract An automated synthesis robot was constructed by modifying an open source 3D printing platform. The resulting automated system
- by the method described, opening possibilities for the sharing of validated synthetic ‘programs’ which can run on similar low cost, user-constructed robotic platforms towards an ‘open-source’ regime in the area of chemical synthesis. Keywords: 3D printing; digitising chemistry; ibuprofen; laboratory
- robotics; open source; reaction ware; Introduction The rapid expansion of 3D-printing technologies in recent decades has been one of the most promising developments in the fields of science and engineering [1]. This technology, along with the open-source ethos and large, committed user and developer base
Integration of enabling methods for the automated flow preparation of piperazine-2-carboxamide
Beilstein J. Org. Chem. 2014, 10, 641–652, doi:10.3762/bjoc.10.56

- Abstract Here we describe the use of a new open-source software package and a Raspberry Pi® computer for the simultaneous control of multiple flow chemistry devices and its application to a machine-assisted, multi-step flow preparation of pyrazine-2-carboxamide – a component of Rifater®, used in the
- of tuberculosis – and its reduced derivative (R,S)-piperazine-2-carboxamide has been demonstrated, using a new open-source software platform for the simultaneous control of multiple devices. The protocol developed here represents a valid example of how these technologies can be used to implement
Mild and efficient cyanuric chloride catalyzed Pictet–Spengler reaction
Beilstein J. Org. Chem. 2013, 9, 1235–1242, doi:10.3762/bjoc.9.140
- a nitrogen atmosphere. Supporting Information Supporting Information File 46: Analytical data and copies of 1H and 13C NMR of 3a, 3c, 3h and 8d. Acknowledgements The authors thank the Open Source Drug Discovery (OSDD) program of CSIR, New Delhi for funding as well as the Department of
Camera-enabled techniques for organic synthesis
Beilstein J. Org. Chem. 2013, 9, 1051–1072, doi:10.3762/bjoc.9.118
- environment. However, the availability of powerful open-source image processing software such as the Python Imaging Library [68] (PIL, an image processing library for the Python [69] programming language) and OpenCV [70] (a C++ library for creating real-time computer vision applications with bindings for the
- the PySerial software library [94], which allows communication via the serial port. This highlights the utility of these open-source resources for the rapid development of new systems. The resulting steady state allowed continuous liquid–liquid separation for a number of days. The first reported use
- 255, and so the threshold of 100 here corresponds to approximately 40% red character. This is sufficient to pick out the region corresponding to the red liquid in the images. (a) Arduino [73][75], a flexible open-source platform for rapidly prototyping electronic applications. (b) Raspberry Pi [74][76
The use of glycoinformatics in glycochemistry
Beilstein J. Org. Chem. 2012, 8, 915–929, doi:10.3762/bjoc.8.104

- , it was developed as an open-source project. Therefore, newly funded projects such as UniCarb-DB [37] or the latest version of GlycoBase (Dublin) [25], which stores HPLC data, are able to make use of the EUROCarbDB source code and, thus, the software does not need to be rewritten. The CFG databases
- disconnected islands. Furthermore, funding for the maintenance of existing databases is required to keep useful resources up-to-date, rather than only funding new projects. The open-source idea can also help to partly overcome this dilemma. If the data and source codes that have been developed in a project are





































































































































































